| CARVIEW |
An official website of the United States government
The .gov means it’s official.
Federal government websites often end in .gov or .mil. Before
sharing sensitive information, make sure you’re on a federal
government site.
The site is secure.
The https:// ensures that you are connecting to the
official website and that any information you provide is encrypted
and transmitted securely.
Save citation to file
Email citation
Add to Collections
Add to My Bibliography
Your saved search
Create a file for external citation management software
Your RSS Feed
Semantic Disease Gene Embeddings (SmuDGE): phenotype-based disease gene prioritization without phenotypes
Affiliation
- 1 Computer, Electrical and Mathematical Sciences and Engineering Division, Computational Bioscience Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia.
- PMID: 30423077
- PMCID: PMC6129260
- DOI: 10.1093/bioinformatics/bty559
Semantic Disease Gene Embeddings (SmuDGE): phenotype-based disease gene prioritization without phenotypes
Authors
Affiliation
- 1 Computer, Electrical and Mathematical Sciences and Engineering Division, Computational Bioscience Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia.
- PMID: 30423077
- PMCID: PMC6129260
- DOI: 10.1093/bioinformatics/bty559
Abstract
Motivation: In the past years, several methods have been developed to incorporate information about phenotypes into computational disease gene prioritization methods. These methods commonly compute the similarity between a disease's (or patient's) phenotypes and a database of gene-to-phenotype associations to find the phenotypically most similar match. A key limitation of these methods is their reliance on knowledge about phenotypes associated with particular genes which is highly incomplete in humans as well as in many model organisms such as the mouse.
Results: We developed SmuDGE, a method that uses feature learning to generate vector-based representations of phenotypes associated with an entity. SmuDGE can be used as a trainable semantic similarity measure to compare two sets of phenotypes (such as between a disease and gene, or a disease and patient). More importantly, SmuDGE can generate phenotype representations for entities that are only indirectly associated with phenotypes through an interaction network; for this purpose, SmuDGE exploits background knowledge in interaction networks comprised of multiple types of interactions. We demonstrate that SmuDGE can match or outperform semantic similarity in phenotype-based disease gene prioritization, and furthermore significantly extends the coverage of phenotype-based methods to all genes in a connected interaction network.
Availability and implementation: https://github.com/bio-ontology-research-group/SmuDGE.
Figures

Fig. 1.
Our knowledge graph consists of…
Fig. 1.
Our knowledge graph consists of gene–phenotype associations (encoded using either HPO or MP),…

Fig. 2.
Overview over SmuDGE and its…
Fig. 2.
Overview over SmuDGE and its applications. ( a ) On the left side…

Fig. 3.
ROC curves for predicting gene–disease…
Fig. 3.
ROC curves for predicting gene–disease associations using cosine similarity between SmuDGE’s P-Vecs and…

Fig. 4.
Comparision of ROC curves for…
Fig. 4.
Comparision of ROC curves for predicting gene–disease associations based on mouse phenotypes using…
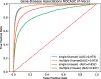
Fig. 5.
ROC curves for predicting gene–disease…
Fig. 5.
ROC curves for predicting gene–disease associations for diseases with a single or multiple…

Fig. 6.
ROC cuves for predicting gene–disease…
Fig. 6.
ROC cuves for predicting gene–disease associations for diseases with a single or multiple…
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
NCBI Literature Resources
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.


