| CARVIEW |
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
Collection
Encode
Access the collected papers by exploring the thematic threads that run through them, with topics such as DNA methylation, RNA or machine learning.

Threads
-
-
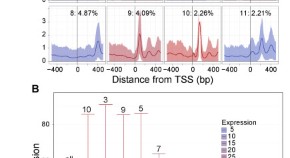
2 Chromatin patterns at transcription factor binding sites
Both chromatin accessibility and histone modifications have different patterns around different transcription factor binding sites
Thread Nature -
3 Characterization of intergenic regions and gene definition
The prevalence and analysis of ENCODE data are changing the definition and characterization of intergenic and genic regions
Thread Nature -
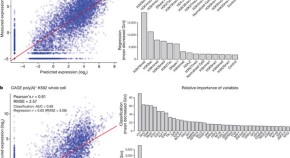
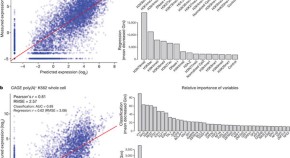
4 RNA and chromatin modification patterns around promoters
Patterns of gene expression can be modelled using histone modifications and transcription factor binding at promoters
Thread Nature -
5 Epigenetic regulation of RNA processing
Many novel and previously known non-coding RNA species are characterized in ENCODE
Thread Nature -

6 Non-coding RNA characterization
Many novel and previously known non-coding RNA species are characterized in ENCODE
Thread Nature -
7 DNA methylation
ENCODE analysis identifies dynamic DNA methylation patterns and relationships to regulatory elements
Thread Nature -
8 Enhancer discovery and characterization
ENCODE has developed methods to discover enhancers, and characterized them using comparisons with other data sets and by molecular biology experiments
Thread Nature -
9 Three-dimensional connections across the genome
ENCODE mapped long-range looping interactions between functional elements and genes, placing them in a three-dimensional context to reveal their functional relationships
Thread Nature -

10 Characterization of network topology
ENCODE data analysis helps to describe the various types of regulatory "wiring" implicit in the genome
Thread Nature -
11 Machine learning approaches to genomics
ENCODE has applied machine learning approaches to enable integration and exploration of large and diverse data
Thread Nature -

12 Impact of functional information on understanding variation
ENCODE provides an initial interpretation of many human variants and plausible leads for the role of many variants identified in genome-wide association studies
Thread Nature -
13 Impact of evolutionary selection on functional regions
The imprint of evolutionary selection on ENCODE regulatory elements is manifested between species and within human populations.
Thread Nature
News and Comment
-

ENCODE: The human encyclopaedia
First they sequenced it. Now they have surveyed its hinterlands. But no one knows how much more information the human genome holds, or when to stop looking for it.
- Brendan Maher
News Feature Nature -

Lessons for big-data projects
To be successful, consortia need clear management, codes of conduct and participants who are committed to working for the common good, says ENCODE lead analysis coordinator Ewan Birney.
- Ewan Birney
Comment Nature -

ENCODE explained
The Encyclopedia of DNA Elements (ENCODE) project dishes up a hearty banquet of data that illuminate the roles of the functional elements of the human genome. Here, six scientists describe the project and discuss how the data are influencing research directions across many fields. See Articles p.57, p.75, p.83, p.91, p.101 & Letter p.109
- Joseph R. Ecker
- Wendy A. Bickmore
- Eran Segal
News & Views Forum Nature -

Collaborations: A single cog in a complex machine
If they remain vigilant, early-career researchers can reap benefits from taking part in big international projects.
- Sarah Kellogg
Feature Nature
Research papers in Nature
-
An integrated encyclopedia of DNA elements in the human genome
This overview of the ENCODE project outlines the data accumulated so far, revealing that 80% of the human genome now has at least one biochemical function assigned to it; the newly identified functional elements should aid the interpretation of results of genome-wide association studies, as many correspond to sites of association with human disease.
- The ENCODE Project Consortium
-

Landscape of transcription in human cells
A description is given of the ENCODE effort to provide a complete catalogue of primary and processed RNAs found either in specific subcellular compartments or throughout the cell, revealing that three-quarters of the human genome can be transcribed, and providing a wealth of information on the range and levels of expression, localization, processing fates and modifications of known and previously unannotated RNAs.
- Sarah Djebali
- Carrie A. Davis
- Thomas R. Gingeras
-

The accessible chromatin landscape of the human genome
An extensive map of human DNase I hypersensitive sites, markers of regulatory DNA, in 125 diverse cell and tissue types is described; integration of this information with other ENCODE-generated data sets identifies new relationships between chromatin accessibility, transcription, DNA methylation and regulatory factor occupancy patterns.
- Robert E. Thurman
- Eric Rynes
- John A. Stamatoyannopoulos
-
An expansive human regulatory lexicon encoded in transcription factor footprints
DNase I footprinting in 41 cell and tissue types reveals millions of short sequence elements encoding an expansive repertoire of conserved recognition sequences for DNA-binding proteins.
- Shane Neph
- Jeff Vierstra
- John A. Stamatoyannopoulos
-

Architecture of the human regulatory network derived from ENCODE data
A description is given of the ENCODE consortium’s efforts to examine the principles of human transcriptional regulatory networks; the results are integrated with other genomic information to form a hierarchical meta-network where different levels have distinct properties.
- Mark B. Gerstein
- Anshul Kundaje
- Michael Snyder
-
The long-range interaction landscape of gene promoters
Chromosome conformation capture carbon copy (5C) is used to look at the relationships between functional elements and distal target genes in 1% of the human genome in three dimensions; the study describes numerous long-range interactions between promoters and distal sites that include elements resembling enhancers, promoters and CTCF-bound sites, their genomic distribution and complex interactions.
- Amartya Sanyal
- Bryan R. Lajoie
- Job Dekker


