| CARVIEW |
The RNA Underworld
Novelty in the polyadenylation machinery
June 7, 2020Late last fall, I published a short review in WIRES RNA that discussed some curious findings coming out of the growing community of plant scientists whose research touches on mRNA polyadenylation. When we think about the polyadenylation machinery, it is reflexive to consider that the core subunits (the CPSF, CstF, CFIm, and CFIIm subunits) should be essential. Indeed, this is the case in yeast and mammals, as far as one can tell. It is thus very surprising that Arabidopsis is able to grow (sometimes, with almost imperceptible phenotypes) in the absence of several supposedly core subunits. The list of dispensable proteins in plants includes CPSF30, FIP1, CstF77, and CstF64.
 3 Comments |
3 Comments |  Evolution, Polyadenylation, RNA Processing | Tagged: Evolution, mutants, Polyadenylation |
Evolution, Polyadenylation, RNA Processing | Tagged: Evolution, mutants, Polyadenylation |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
What we’ve learned during the blogging hiatus
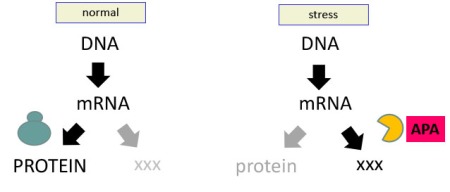
May 25, 2020It’s been seven years or so since I updated this blog. In this post, I hope to summarize a direction my lab has taken in this time. If I am successful, you will see how interesting and exciting this research is, and the new directions we are exploring.
For the better part of 12 years, a main focus of research in my lab has been alternative polyadenylation in plants. I summarized a couple of seminal papers previously – here and here. One of the take-home points of this research was the scope of alternative polyadenylation – how many different poly(A) sites could be used, how usage shifts, and the impacts that shifts in poly(A) site choice have on gene expression. Since 2013, we have published many additional papers on alternative polyadenylation. The three summarized here help to develop a theme that guides current research in my lab.
 2 Comments |
2 Comments |  Polyadenylation, Regulation of gene expression, RNA Processing |
Polyadenylation, Regulation of gene expression, RNA Processing |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
Well, that was a bit of a break
May 24, 2020Hmm… I guess that hiatus was a bit longer than I had anticipated. It’s been more than 7 years, so I guess a few updates are in order. Over the next posts, I will bring this blog up to speed – stuff about my lab, other science-related activities, and odds and ends as befits the history of this blog. Read the rest of this entry »
 Leave a Comment » |
Leave a Comment » |  About me |
About me |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
While in Lexington …
April 7, 2013… for the recently-started Spring Meet at Keeneland (it really is this pretty), I suggest that you find time to attend the 2013 Naff Symposium at the University of Kentucky. This is an annual event put on by the Dept. of Chemistry and centers on aspects of chemistry and molecular biology. This year’s topic is The Origin of Life, and the line-up of speakers is pretty amazing.
 4 Comments |
4 Comments |  About me, Evolution, Origins of Life, Uncategorized |
About me, Evolution, Origins of Life, Uncategorized |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
On the 47th day …
February 17, 201347 days into the new year. 5 grant proposals. What to do to interrupt the tedium?
 3 Comments |
3 Comments |  About me, Odds and ends |
About me, Odds and ends |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
Patenting genes
February 8, 2013With a tip of the hat to Tim Sandefur and The Panda’s Thumb, an interesting discussion at SCOTUSBlog on the the case of Association of Molecular Pathology v. Myriad Genetics, one that involves the issue of the patentability of human genes. Enjoy.
 Leave a Comment » |
Leave a Comment » |  Biotechnology |
Biotechnology |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
How bad …
December 31, 2012… was it? So bad that my mother called me immediately after the game. Not to console me, but to gloat.
(What game? If you have to ask, then …)
 2 Comments |
2 Comments |  About me, Kentucky basketball |
About me, Kentucky basketball |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
You know …
December 29, 2012… you’re in a crazy place when the first 11 articles on the Kentucky.com mobile app are about a basketball game. Not a tournament game. Not even a league game. Just a game that a governor decree must be played.
In case you’re wondering, guilty as charged.
 Leave a Comment » |
Leave a Comment » |  Kentucky basketball |
Kentucky basketball |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
Just how widespread is alternative polyadenylation in plants?
November 14, 2012This is the question I think about a lot, and one I spent a some time on in a recent minireview. The answer is, in a nutshell, very.
One of the things I had to do for this review was try and make sense out of the different approaches that have been described recently for studying alternative poly(A) site choice in plants. One of these – the use of high-throughput sequencing to sequence cDNA tags that query the exact mRNA-poly(A) junction – has been discussed previously, in a general sense and in terms of a study of poly(A) site choice in plants. In the latter study, it was determined that about 70% of plant genes possess at least two poly(A) sites.
 1 Comment |
1 Comment |  About me, Polyadenylation, Regulation of gene expression |
About me, Polyadenylation, Regulation of gene expression |  Permalink
Permalink
 Posted by Arthur Hunt
Posted by Arthur Hunt
-
Recent Posts
Recent Comments
Categories
Biotechnology
Blogroll
My research
-
Join 29 other subscribers
Archives
- July 2020
- June 2020
- May 2020
- April 2013
- February 2013
- December 2012
- November 2012
- May 2012
- April 2012
- March 2012
- December 2011
- October 2011
- August 2011
- July 2011
- June 2011
- March 2011
- February 2011
- December 2010
- November 2010
- October 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- August 2008
- July 2008
- June 2008
- May 2008
Meta
January 2026 S M T W T F S 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 Copyright stuff
The content of this blog is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 United States License.Disclaimer:
The opinions voiced on this blog are entirely those of the author, and should in no way be construed as being representative of his employer or of any other agency or organization.
-
Subscribe
Subscribed
Already have a WordPress.com account? Log in now.