Pairwise Deletion is one method used to handle missing data, especially when estimating correlations or covariances.
Missing Data
Missing data can occur due to various reasons, such as sensor failures, manual entry errors or respondents skipping survey questions. Missing data mechanisms are categorized as:
- MCAR (Missing Completely at Random): The probability of missingness is independent of observed or unobserved data.
- MAR (Missing at Random): The probability of missingness depends on observed data but not unobserved data.
- MNAR (Missing Not at Random): The probability of missingness depends on unobserved data.
Challenges Posed by Missing Data
Pairwise Deletion
Pairwise deletion is a technique that evaluates each pair of variables based only on cases (rows) that have non-missing values for that pair. In simpler terms, it computes statistics such as correlations or covariances using the maximum number of available data points for each pair.
This approach contrasts with listwise deletion, which removes entire rows from the dataset if any value is missing.
To understand pairwise deletion, let us consider an example of calculating the correlation coefficient 𝑟 between two variables 𝑋 and 𝑌.
r = \frac{\sum_{i=1}^{n}(x_i - \bar{X})(y_i - \bar{Y})}{\sqrt{\sum_{i=1}^{n}(x_i - \bar{X})^2 \cdot \sum_{i=1}^{n}(y_i - \bar{Y})^2}}
Where:
- n: Number of observations (data points) in the dataset.
- xi, yi: Individual values of variables X and Y respectively.
- \bar{X}, \bar{Y}: Mean (average) of X and Y
Pairwise Deletion in Machine Learning
Python
import numpy as np
import pandas as pd
# Sample dataset with missing values
data = {
'X': [1, 2, np.nan, 4, 5],
'Y': [5, np.nan, 2, 4, 3],
'Z': [2, 3, 4, 5, np.nan]
}
df = pd.DataFrame(data)
def pairwise_correlation(df):
"""Compute pairwise correlations."""
correlations = {}
for col1 in df.columns:
for col2 in df.columns:
if col1 != col2:
# Drop rows where either value is missing
valid_data = df[[col1, col2]].dropna()
if not valid_data.empty:
corr = valid_data.corr().iloc[0, 1]
correlations[f'{col1}-{col2}'] = corr
return correlations
correlations = pairwise_correlation(df)
print("Pairwise Correlations:")
print(correlations)
OutputPairwise Correlations:
{'X-Y': np.float64(-0.9607689228305226), 'X-Z': np.float64(1.0), 'Y-X': np.float64(-0.9607689228305226), 'Y-Z': np.float64(-0.4999999999999999), 'Z-X': np.float64(1.0), 'Z-Y': n... Pairwise Deletion in Deep Learning
In deep learning, handling missing data typically involves imputation techniques rather than deletion. However, pairwise deletion might still be used during preprocessing for correlation analysis.
Visualizing Pairwise Correlations with a Heatmap
Python
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# Step 1: Create a sample DataFrame (Replace this with your actual dataset)
data = {
"A": [1, 2, 3, 4, 5],
"B": [2, 3, 4, 5, 6],
"C": [5, 4, 3, 2, 1],
"D": [3, 3, 3, 3, 3]
}
df = pd.DataFrame(data) # Define the DataFrame
# Step 2: Compute the pairwise correlation matrix
pairwise_corr = df.corr(min_periods=1) # Ensure there are enough non-NaN values
# Step 3: Create a heatmap using Seaborn
sns.heatmap(pairwise_corr, annot=True, cmap="coolwarm")
# Step 4: Customize and display the plot
plt.title("Pairwise Correlation Heatmap")
plt.show()
Output:
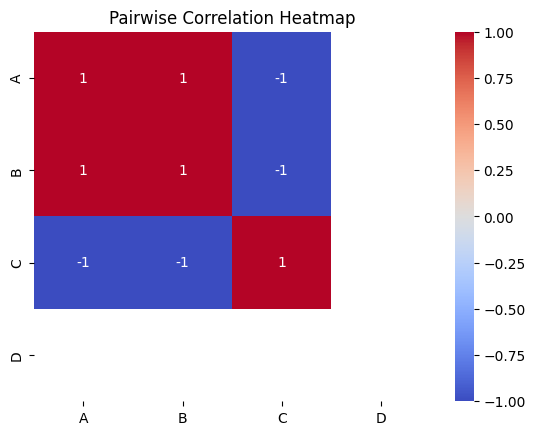 Pairwise Correlation Heatmap
Pairwise Correlation HeatmapAlternatives to Pairwise Deletion
While pairwise deletion has its merits, it is not always the best method. Here are some alternatives:
- Listwise Deletion: Removes rows with any missing values.
- Mean/Median Imputation: Replaces missing values with the mean or median of the feature.
- Multiple Imputation: Generates multiple plausible datasets and averages results.
- Machine Learning Imputation: Uses models like k-Nearest Neighbors or MICE (Multiple Imputation by Chained Equations).
- Matrix Factorization: Fills missing values by leveraging low-rank structures in the data.
Advantages of Pairwise Deletion
- Maximizes Data Usage: Retains more data compared to listwise deletion.
- Simple to Implement: Pairwise deletion is computationally straightforward for small datasets.
- Preserves Relationships: Useful for specific analyses like correlation estimation.
Disadvantages of Pairwise Deletion
- Inconsistent Sample Sizes: Each pair of variables may use a different subset of data, leading to challenges in comparing results.
- Potential Bias: If the data is not MCAR, pairwise deletion may introduce bias.
- Not Suitable for Complex Models: For regression or machine learning models, inconsistent sample sizes can complicate training.
